How To Setup A Smart Daq To Upload Live Data To A Repository
About the project
This project was for prototyping smart DAQ devices able to do data measurements at a research laboratory and do automatic experimental data upload to a repository at Harvard University 's dataverse
Items used in this project
Hand tools and fabrication machines
Story
The other day I found a new way of doing scientific research science spite research data repositories exist for quite some time now. And this was not by chance, after one year of upgrading my hardware electronics design skills, and now have more than 20 PCB design projects available on GitHub ready to help me do real LIVE & publishable science.
_________________________________________________________________________
this article is being used to write a MethodX paper suitable for publication in a peer-reviewed journal. Updates to this article will continue here and here as I write the paper in parallel
_________________________________________________________________________
The novelty to me is the possibility of programming some firmware piece of code into any of my custom designed, prototyped, and built smart data acquisition devices, named #LDAD (Live Data Acquisition Device) to upload automatically to a scientific data repository, all experimental data being collected on any of my experimental setups. For instance Harvard University's #Dataverse.
Video above shows the final result where the smart DAQ is installed on top of a self-sensing smart asphalt specimen ready for data measurements collection
Moreover, adding to this huge advantage in designing an experiment, is possible to upload #LIVE data to such data repositories. This means I can finally do scientific collaboration and cooperation with anyone working on the same subject and on any country on the planet. I'm no longer bound to limitations of proximity, language, or secrecy. The advantages do not end here, this way of doing experimental data collection gives a researcher the freedom, and time, by minimizing the need for schedule synchronization among fellow researchers collaborating, in particular, useless remote meetings over Microsoft #Teams or #Zoom or even other, now redundant, socializing tasks.
List of data repositories with an API
- Zenodo: ReST API; Open Source;
- OSF : API ; Open Source;
- Harvard's Dataverse: API; Open Source;
- DRYAD: NO;
- Mendeley: API;
- Data in Brief: NO;
- Scientific Data: NO;
One mandatory requirement is for the reader to be knowledgeable about how to work in open science environments, in particular how to use Open Data, Open Source, and Open Hardware electronics. There's a short introductory video on YouTube about "Open Data & Data Sharing Platforms" in science by Shady Attia, a prof. at the University of Liège, which I recommend watching before reading further. Another requirement is for the reader to be experienced in how to correct errors, for instance by the level of severity, on his/her LIVE public scientific research. An open AGILE way of doing science, with constant updates and feedback made publicly and in real-time. These are mandatory requirements for all looking to do science in a live and real-time manner and take advantage of all that live open environments have to offer. In regards to safety & health in science. In regards to Intellectual property. In regards to remote collaboration and cooperation among scientific researchers.
unedited video with the very first smart DAQ components assembly into the EuroCircuits PCB.
For this short tutorial, I'll be using Harvard University's #Dataverse #API with a simple hack. Anyone can find the API documentation here. The piece of API of interest is this one, replace of dataset file on a repository:
export API_TOKEN=xxxxxxxx-xxxx-xxxx-xxxx-xxxxxxxxxxxx export SERVER_URL=https://demo.dataverse.org export ID=24 curl -H "X-Dataverse-key:$API_TOKEN" -X POST -F 'file=@file.extension' -F 'jsonData={json}' $SERVER_URL/api/files/$ID/replace
My 12/24bit LDAD DAQ devices use Tensilica LX7 dual-core MCUs form Expressif and are compatible with Arduino Studio. Below is the piece of code to send locally collected sensor data, as a file, to a dataverse repository using their API:
- #include <SPI.h>
- #include <string.h>
- #include “utility/debug.h”
- #include<stdlib.h>
- #include <SD.h>
- // WiFi network (change with your settings !)
- #define WLAN_SSID “mywirelessnetwork”
- #define WLAN_PASS “mypassword”
- # define WLAN_SECURITY WLAN_SEC_WPA2
- WiFiClient client;
- int i = 0;
- int keyIndex = 1;
- int port = 80; //Server Port
- uint32_t MANip = 0;
- //************* == LOOP == ************************* unsigned long lastMillisSensors=0; unsigned long lastMillisWIFI=0; bool scheduleWait = false; int waitTimeSensors=0; int waitTimeWIFI=0; //*********************** == Core 2 == ************************ void loop1() { // WIFI ********************** digitalWrite(ledBlue, LOW); connect2WIFInetowrk(); digitalWrite(ledBlue, HIGH); digitalWrite(ledGreen, LOW); delay(1000); if (UPLOAD_DATASET_DELTA_TIME < ( millis() - LAST_DATASET_UPLOAD )){ LAST_DATASET_UPLOAD= millis(); digitalWrite(ledBlue, HIGH); digitalWrite(ledGreen, LOW); delay(1000); mserial.printStrln(""); mserial.printStrln("Upload Data to the Dataverse..."); while (datasetFileIsBusySaveData){ mserial.printStr("busy S "); delay(500); } xSemaphoreTake(MemLockSemaphoreDatasetFileAccess, portMAX_DELAY); // enter critical section datasetFileIsBusyUploadData=true; xSemaphoreGive(MemLockSemaphoreDatasetFileAccess); // exit critical section mserial.printStrln(" get dataset metadata..."); getDatasetMetadata(); UploadToDataverse(0); xSemaphoreTake(MemLockSemaphoreDatasetFileAccess, portMAX_DELAY); // enter critical section datasetFileIsBusyUploadData=false; xSemaphoreGive(MemLockSemaphoreDatasetFileAccess); // exit critical section digitalWrite(ledBlue, HIGH); digitalWrite(ledGreen, LOW); delay(1000); xSemaphoreTake(MemLockSemaphoreCore2, portMAX_DELAY); // enter critical section waitTimeWIFI=0; xSemaphoreGive(MemLockSemaphoreCore2); // exit critical section } if (millis()-lastMillisWIFI > 60000){ xSemaphoreTake(MemLockSemaphoreCore2, portMAX_DELAY); // enter critical section waitTimeWIFI++; lastMillisWIFI=millis(); xSemaphoreGive(MemLockSemaphoreCore2); // exit critical section } } // ********************* == Core 1 == ****************************** void loop() { if (DO_DATA_MEASURMENTS_DELTA_TIME < ( millis() - LAST_DATA_MEASUREMENTS ) ){ LAST_DATA_MEASUREMENTS=millis(); digitalWrite(ledRed, LOW); digitalWrite(ledGreen, HIGH); delay(1000); mserial.printStrln(""); mserial.printStrln("Reading Specimen electrical response..."); updateInternetTime(); ReadExternalAnalogData(); digitalWrite(ledRed, HIGH); digitalWrite(ledGreen, LOW); delay(1000); mserial.printStrln("Saving collected data...."); if(datasetFileIsBusyUploadData){ mserial.printStr("file is busy...."); } if(measurements_current_pos+1 > MEASUREMENTS_BUFFER_SIZE){ mserial.printStr("[mandatory wait]"); while (datasetFileIsBusyUploadData){ } initSaveDataset(); }else{ // measurements buffer is not full long latency=millis(); long delta= millis()-latency; while (datasetFileIsBusyUploadData || (delta < MAX_LATENCY_ALLOWED)){ delta= millis()-latency; delay(500); } if(datasetFileIsBusyUploadData){ measurements_current_pos++; mserial.printStr("skipping file save."); }else{ initSaveDataset(); } } scheduleWait=false; xSemaphoreTake(MemLockSemaphoreCore1, portMAX_DELAY); // enter critical section waitTimeSensors=0; xSemaphoreGive(MemLockSemaphoreCore1); // exit critical section } if (scheduleWait){ mserial.printStrln(""); mserial.printStrln("Waiting (schedule) .."); } if (millis()-lastMillisSensors > 60000){ xSemaphoreTake(MemLockSemaphoreCore1, portMAX_DELAY); // enter critical section lastMillisSensors=millis(); mserial.printStrln("Sensor Acq. in " + String((DO_DATA_MEASURMENTS_DELTA_TIME/60000)-waitTimeSensors) + " min </> Upload Experimental Data to a Dataverse Repository in " + String((UPLOAD_DATASET_DELTA_TIME/60000)-waitTimeWIFI) + " min"); waitTimeSensors++; xSemaphoreGive(MemLockSemaphoreCore1); // exit critical section } } void initSaveDataset(){ mserial.printStrln("starting"); digitalWrite(ledRed, HIGH); digitalWrite(ledGreen, LOW); // SAVE DATA MEASUREMENTS **** xSemaphoreTake(MemLockSemaphoreDatasetFileAccess, portMAX_DELAY); // enter critical section datasetFileIsBusySaveData=true; xSemaphoreGive(MemLockSemaphoreDatasetFileAccess); // exit critical section delay(1000); saveDataMeasurements(); xSemaphoreTake(MemLockSemaphoreDatasetFileAccess, portMAX_DELAY); // enter critical section datasetFileIsBusySaveData=false; xSemaphoreGive(MemLockSemaphoreDatasetFileAccess); // exit critical section digitalWrite(ledRed, LOW); digitalWrite(ledGreen, HIGH); }
- void UploadToDataverse(byte counter) { if(counter>5) return; //Check WiFi connection status if(WiFi.status() != WL_CONNECTED){ mserial.printStrln("WiFi Disconnected"); if (connect2WIFInetowrk()){ UploadToDataverse(i+1); } } bool uploadStatusNotOK=true; if(datasetInfoJson["data"].containsKey("id")){ if(datasetInfoJson["data"]["id"] !=""){ String rawResponse = GetInfoFromDataverse("/api/datasets/"+ datasetInfoJson["data"]["id"] +"/locks"); const size_t capacity =2*rawResponse.length() + JSON_ARRAY_SIZE(1) + 7*JSON_OBJECT_SIZE(1); DynamicJsonDocument datasetLocksJson(capacity); // Parse JSON object DeserializationError error = deserializeJson(datasetLocksJson, rawResponse); if (error) { mserial.printStr("unable to retrive dataset lock status. Upload not possible. ERR: "+error.f_str()); //mserial.printStrln(rawResponse); return; }else{ String stat = datasetInfoJson["status"]; if(datasetInfoJson.containsKey("lockType")){ String locktype = datasetInfoJson["data"]["lockType"]; mserial.printStrln("There is a Lock on the dataset: "+ locktype); mserial.printStrln("Upload of most recent data is not possible without removal of the lock."); // Do unlocking }else{ mserial.printStrln("The dataset is unlocked. Upload possible."); uploadStatusNotOK=false; } } }else{ mserial.printStrln("dataset ID is empty. Upload not possible. "); } }else{ mserial.printStrln("dataset metadata not loaded. Upload not possible. "); } if(uploadStatusNotOK){ return; } // Open the dataset file and prepare for binary upload File datasetFile = FFat.open("/"+EXPERIMENTAL_DATA_FILENAME, FILE_READ); if (!datasetFile){ mserial.printStrln("Dataset file not found"); return; } String boundary = "7MA4YWxkTrZu0gW"; String contentType = "text/csv"; DATASET_REPOSITORY_URL = "/api/datasets/:persistentId/add?persistentId="+PERSISTENT_ID; String datasetFileName = datasetFile.name(); String datasetFileSize = String(datasetFile.size()); mserial.printStrln("Dataset File Details:"); mserial.printStrln("Filename:" + datasetFileName); mserial.printStrln("size (bytes): "+ datasetFileSize); mserial.printStrln(""); int str_len = SERVER_URL.length() + 1; // Length (with one extra character for the null terminator) char SERVER_URL_char [str_len]; // Prepare the character array (the buffer) SERVER_URL.toCharArray(SERVER_URL_char, str_len); // Copy it over client.stop(); client.setCACert(HARVARD_ROOT_CA_RSA_SHA1); if (!client.connect(SERVER_URL_char, SERVER_PORT)) { mserial.printStrln("Cloud server URL connection FAILED!"); mserial.printStrln(SERVER_URL_char); int server_status = client.connected(); mserial.printStrln("Server status code: " + String(server_status)); return; } mserial.printStrln("Connected to the dataverse of Harvard University"); mserial.printStrln(""); mserial.printStr("Requesting URL: " + DATASET_REPOSITORY_URL); // Make a HTTP request and add HTTP headers String postHeader = "POST " + DATASET_REPOSITORY_URL + " HTTP/1.1rn"; postHeader += "Host: " + SERVER_URL + ":" + String(SERVER_PORT) + "rn"; postHeader += "X-Dataverse-key: " + API_TOKEN + "rn"; postHeader += "Content-Type: multipart/form-data; boundary=" + boundary + "rn"; postHeader += "Accept: text/html,application/xhtml+xml,application/xml,application/json;q=0.9,*/*;q=0.8rn"; postHeader += "Accept-Encoding: gzip,deflatern"; postHeader += "Accept-Charset: ISO-8859-1,utf-8;q=0.7,*;q=0.7rn"; postHeader += "User-Agent: AeonLabs LDAD Smart DAQ devicern"; postHeader += "Keep-Alive: 300rn"; postHeader += "Connection: keep-alivern"; postHeader += "Accept-Language: en-usrn"; // jsonData header String jsonData = "{"description":"LIVE Experimental data upload from LDAD Smart 12bit DAQ ","categories":["Data"], "restrict":"false", "tabIngest":"false"}"; String jsonDataHeader = "--" + boundary + "rn"; jsonDataHeader += "Content-Disposition: form-data; name="jsonData"rnrn"; jsonDataHeader += jsonData+"rn"; // dataset header String datasetHead = "--" + boundary + "rn"; datasetHead += "Content-Disposition: form-data; name="file"; filename="" + datasetFileName + ""rn"; datasetHead += "Content-Type: " + contentType + "rnrn"; // request tail String tail = "rn--" + boundary + "--rnrn"; // content length int contentLength = jsonDataHeader.length() + datasetHead.length() + datasetFile.size() + tail.length(); postHeader += "Content-Length: " + String(contentLength, DEC) + "nn"; // send post header int postHeader_len=postHeader.length() + 1; char charBuf0[postHeader_len]; postHeader.toCharArray(charBuf0, postHeader_len); client.print(charBuf0); mserial.printStr(charBuf0); // send key header char charBufKey[jsonDataHeader.length() + 1]; jsonDataHeader.toCharArray(charBufKey, jsonDataHeader.length() + 1); client.print(charBufKey); mserial.printStr(charBufKey); // send request buffer char charBuf1[datasetHead.length() + 1]; datasetHead.toCharArray(charBuf1, datasetHead.length() + 1); client.print(charBuf1); mserial.printStr(charBuf1); // create buffer const int bufSize = 2048; byte clientBuf[bufSize]; int clientCount = 0; while (datasetFile.available()) { clientBuf[clientCount] = datasetFile.read(); clientCount++; if (clientCount > (bufSize - 1)) { client.write((const uint8_t *)clientBuf, bufSize); clientCount = 0; } } datasetFile.close(); if (clientCount > 0) { client.write((const uint8_t *)clientBuf, clientCount); mserial.printStrln("[binary data]"); } // send tail char charBuf3[tail.length() + 1]; tail.toCharArray(charBuf3, tail.length() + 1); client.print(charBuf3); mserial.printStr(charBuf3); // Read all the lines on reply back from server and print them to mserial mserial.printStrln(""); mserial.printStrln("Response Headers:"); String responseHeaders = ""; while (client.connected()) { // mserial.printStrln("while client connected"); responseHeaders = client.readStringUntil('n'); mserial.printStrln(responseHeaders); if (responseHeaders == "r") { mserial.printStrln("====== end of headers ======"); break; } } String responseContent = client.readStringUntil('n'); mserial.printStrln("Harvard University's Dataverse reply was:"); mserial.printStrln("=========="); mserial.printStrln(responseContent); mserial.printStrln("=========="); mserial.printStrln("closing connection"); client.stop(); }
(see full code on Github here)
The piece of code above only handles sending collected data, stored in a local file, to a dataverse repository. To store collected data locally on the smart DAQ device, data is needed first to be saved on the local file system SPIFS. The reader can find a good tutorial on Tutorials Point, with code examples on how to use the SPIFS library. Below is part of the code needed that focus on saving the actual sensor data.
- // *********************************************************
- // FS FAT File Management // ********************************************************* // create new CSV ; delimeted dataset file bool initializeDataMeasurementsFile(){ if (FFat.exists("/" + EXPERIMENTAL_DATA_FILENAME)){ return true; } // create new CSV ; delimeted dataset file EXPERIMENTAL_DATA_FILE = FFat.open("/" + EXPERIMENTAL_DATA_FILENAME,"w"); if (EXPERIMENTAL_DATA_FILE){ mserial.printStr("Creating a new dataset CSV file and adding header ..."); String DataHeader[NUM_DATA_READINGS]; DataHeader[0]="Date&Time"; DataHeader[1]="ANALOG RAW (0-4095)"; DataHeader[2]="Vref (Volt)"; DataHeader[3]="V (Volt)"; DataHeader[4]="R (Ohm)"; DataHeader[5]="SHT TEMP (*C)"; DataHeader[6]="SHT Humidity (%)"; DataHeader[7]="Accel X"; DataHeader[8]="Accel Y"; DataHeader[9]="Accel Z"; DataHeader[10]="Gyro X"; DataHeader[11]="Gyro Y"; DataHeader[12]="Gyro Z"; DataHeader[13]="LSM6DS3 Temp (*C)"; DataHeader[14]="Bus non-errors"; DataHeader[15]="Bus Erros"; DataHeader[16]="Validation"; DataHeader[17]="CHIP ID"; String lineRowOfData=""; for (int j = 0; j < NUM_DATA_READINGS; j++) { lineRowOfData= lineRowOfData + DataHeader[j] +";"; } EXPERIMENTAL_DATA_FILE.println(lineRowOfData); EXPERIMENTAL_DATA_FILE.close(); mserial.printStrln("header added to the dataset file."); }else{ mserial.printStrln("Error creating file(2): " + EXPERIMENTAL_DATA_FILENAME); return false; } mserial.printStrln(""); return true; } // ********************************************************* bool saveDataMeasurements(){ EXPERIMENTAL_DATA_FILE = FFat.open("/" + EXPERIMENTAL_DATA_FILENAME,"a+"); if (EXPERIMENTAL_DATA_FILE){ mserial.printStrln("Saving data measurements to the dataset file ..."); for (int k = 0; k <= measurements_current_pos; k++) { for (int i = 0; i < NUM_SAMPLES; i++) { String lineRowOfData=""; for (int j = 0; j < NUM_DATA_READINGS; j++) { lineRowOfData= lineRowOfData + measurements[k][i][j] +";"; } lineRowOfData+= macChallengeDataAuthenticity(lineRowOfData) + ";" + CryptoICserialNumber(); mserial.printStrln(lineRowOfData); EXPERIMENTAL_DATA_FILE.println(lineRowOfData); } } EXPERIMENTAL_DATA_FILE.close(); delay(500); mserial.printStrln("collected data measurements stored in the dataset CSV file.("+ EXPERIMENTAL_DATA_FILENAME +")"); measurements_current_pos=0; return true; }else{ mserial.printStrln("Error creating CSV dataset file(3): " + EXPERIMENTAL_DATA_FILENAME); return false; } }
(see full code on Github here)
As I stated at the beginning of this short tutorial, this is kind of a #hack to dataverse's API. And the hack is on recurrently uploading a replacement dataset file to the repository. To this date, the API lacks the functionality of uploading raw sensor data to be appended by the dataverse system to an existing file hosted in a repository, the code above does it locally on the smart DAQ, saving collected sensor data and storing it in a local file. This means the smart DAQ needs to have enough storage space to save all experimental data collected during the experimental campaign. Another important consideration to have is the number of times the local file, stored in the smart DAQ, is uploaded to a dataverse repository. Too many times, in a short period of time, may result in a temporary loss of access from the dataverse server. Too few times, other researchers accessing data remotely are kept waiting and are unable to work on experimental data in a LIVE manner. This needs to be inquired to the host of the data repository for an accurate number of file uploads allowed.
One Final remark, to add the following. The best API for this kind of scientific research needs to be capable of LIVE data storage and accept LIVE raw sensor data. For instance, accept:
- an inline string with sensor data and append it to an existing file CSV file on a repository.
- sensor data plus information on where to be added in an excel document file. For instance in a specific column/ row.
- sensor data plus information of table and table fields to be added in an SQLite database file.
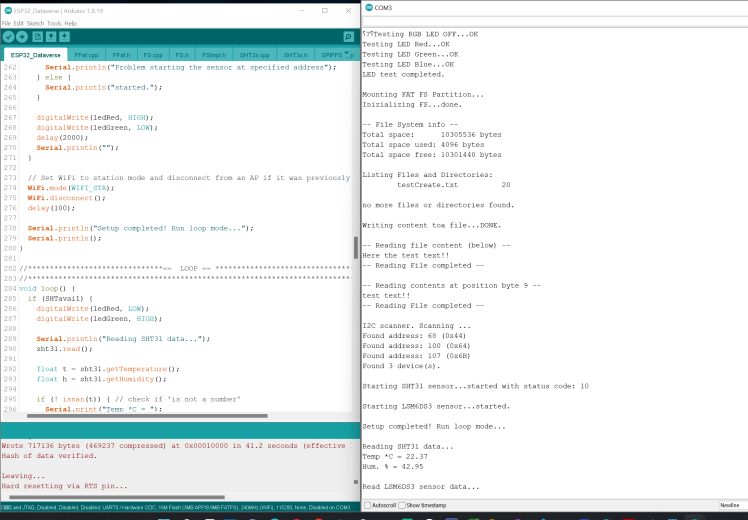
snapshot with a serial output showing PCB onboard sensor testing and working.
In 2022, storing experimental data makes more sense by doing it in an individual file rather than on a common centralized database server. The advantages of individual files, Excel or SQLite, over a common database can be summarized due to the centralized nature of the latter. In a centralized form, is more difficult and more complex to share experimental data contents. It also requires additional software and setup for someone wanting to access it offline. Another advantage over a centralized DB server is the fact individual files can be stored in a censorship-resistant web3, for instance on the Interplanetary File System (IPFS) Network. Accessing data is as easy as ... well opening MS Excel. For cases of Big, huge data records above Excel maximum number of allowed data rows, SQLite database files are well suited and fully compatible with Python or any other programming language with more than enough open source libraries available on GitHub.
CAD, enclosures and custom parts
Code
Credits

Aeon
During my daily work activity I do Research , Prototype and Deployment of Smart AI enabled Technology Solutions. I also Advise and Mentor technology solutions to individuals and organizations (enterprises and institutions). I prototyped and about to publish the 3rd scientific paper on self sensing carbon fiber composite materials for active structural monitoring , and for the past 4 months now, I've been prototyping smart DAC devices to connect such materials to and edge server. The main objective, give life to a structure, by seamlessly integrating any structural element into a remote, live active (or passive) monitoring network with usage of artificial intelligence technologies all together
































Leave your feedback...